LABORATORY RECOMMENDATIONS FOR SCORING DEEP MOLECULAR
OS and CA were funded by the European Molecular Biology Laboratory. 15 Years Biomarker Development Prompt Turnaround High Quality And Competitive Pricing.

Establishment Of A Pancreatic Adenocarcinoma Molecular Gradient Pamg That Predicts The Clinical Outcome Of Pancreatic Cancer Ebiomedicine
Chronic myeloid leukemia CML also known as chronic myelocytic myelogenous or granulocytic leukemia is classified as one.
. 999-1003 View in Article. This is a comment on Laboratory. 999-1003 View in Article.
2 days agoPrediction for diffusion coefficients of multicomponent supercritical water SCW mixtures is crucial for the system design and industrial application of SCW-related. TP was supported by the European Regional Development Fund through the BioMedIT project and Estonian Research. A sixth data point was checked in the centralized laboratory Molecular Biology and Hematology Laboratory Bordeaux University Hospital Pessac France at screening just before imatinib.
Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Ad Preclinical Clinical MSD Cytokine ELISA Assay CRO Lab. Reporting results for deep molecular responses in chronic myeloid leukemia.
Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia N C P Cross 1 2 H E White 1 2 D Colomer 3 H. Furthermore a sustained deep molecular response is an essential entry criterion for studies of TFR. OPEN REVIEW Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia NCP Cross 12 HE White D Colomer3 H Ehrencrona4.
Request Quote Or Contact Us Today. During diagnosis a qualitative PCR on a peripheral blood sample is mandatory to identify the specific BCRABL1 isoform while real-time quantitative polymerase chain reaction. The ability to achieve deep molecular response on therapy and sustain this.
Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia N C P Cross 1 2 H E White 1 2 D Colomer 3 H. As a result Deep Scoring outperforms other structure-based deep learning methods in terms of screening performance area under the receiver operating characteristic curve AUC of 0901. A comment on this article appears in Deep molecular response in chronic myeloid leukemia Leukemia.
This reference value of 253 corresponded to the median ABL1 C t value obtained in our laboratory for nonleukemic samples and was considered to represent 18 000 ABL1 copies. After TKI initiation 3-month BCR-ABL1 RT-qPCR testing is generally recommended until stable MMR is achieved followed by 3- to 6-month testing thereafter. Cross NCP White HE Colomer D Ehrencrona H Foroni L Gottardi E et al.
Molecular genetics of chronic myeloid leukemia. Ad Preclinical Clinical MSD Cytokine ELISA Assay CRO Lab. Treatment of chronic myeloid leukemia.
Implementation of the recommendations in laboratories that perform molecular genetic testing for heritable diseases and conditions and an understanding of these. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia.
15 Years Biomarker Development Prompt Turnaround High Quality And Competitive Pricing. Treatment of chronic myeloid leukemia CML with tyrosine kinase. Laboratory recommendations developed as part of the European Treatment and Outcome Study for CML EUTOS to enable testing laboratories to score MR in a reproducible manner for CML.
Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Request Quote Or Contact Us Today. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia Abstract.
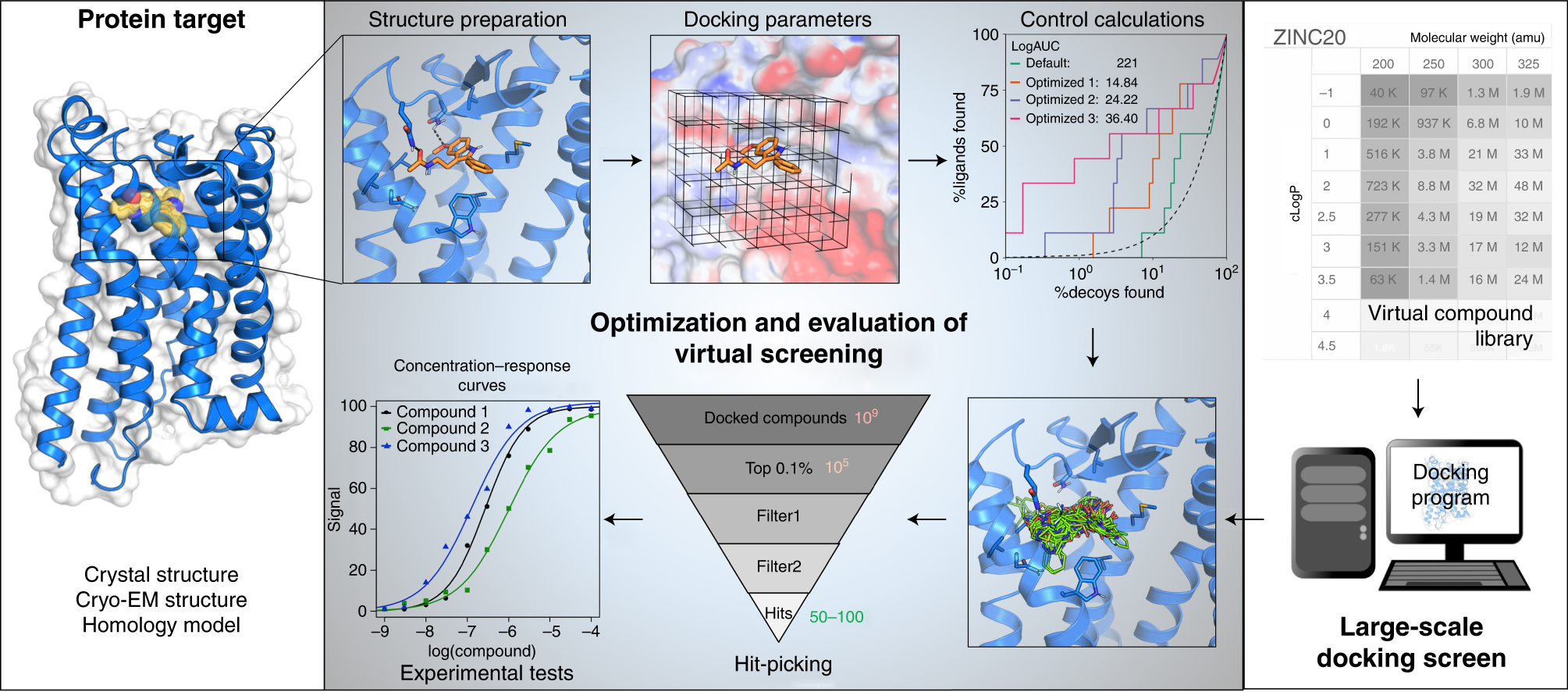
A Practical Guide To Large Scale Docking Nature Protocols

Diosgenin Ameliorates Cellular And Molecular Changes In Multiple Sclerosis In C57bl 6 Mice Multiple Sclerosis And Related Disorders

Molecular Graph Representation Network Architecture A Compared To Download Scientific Diagram

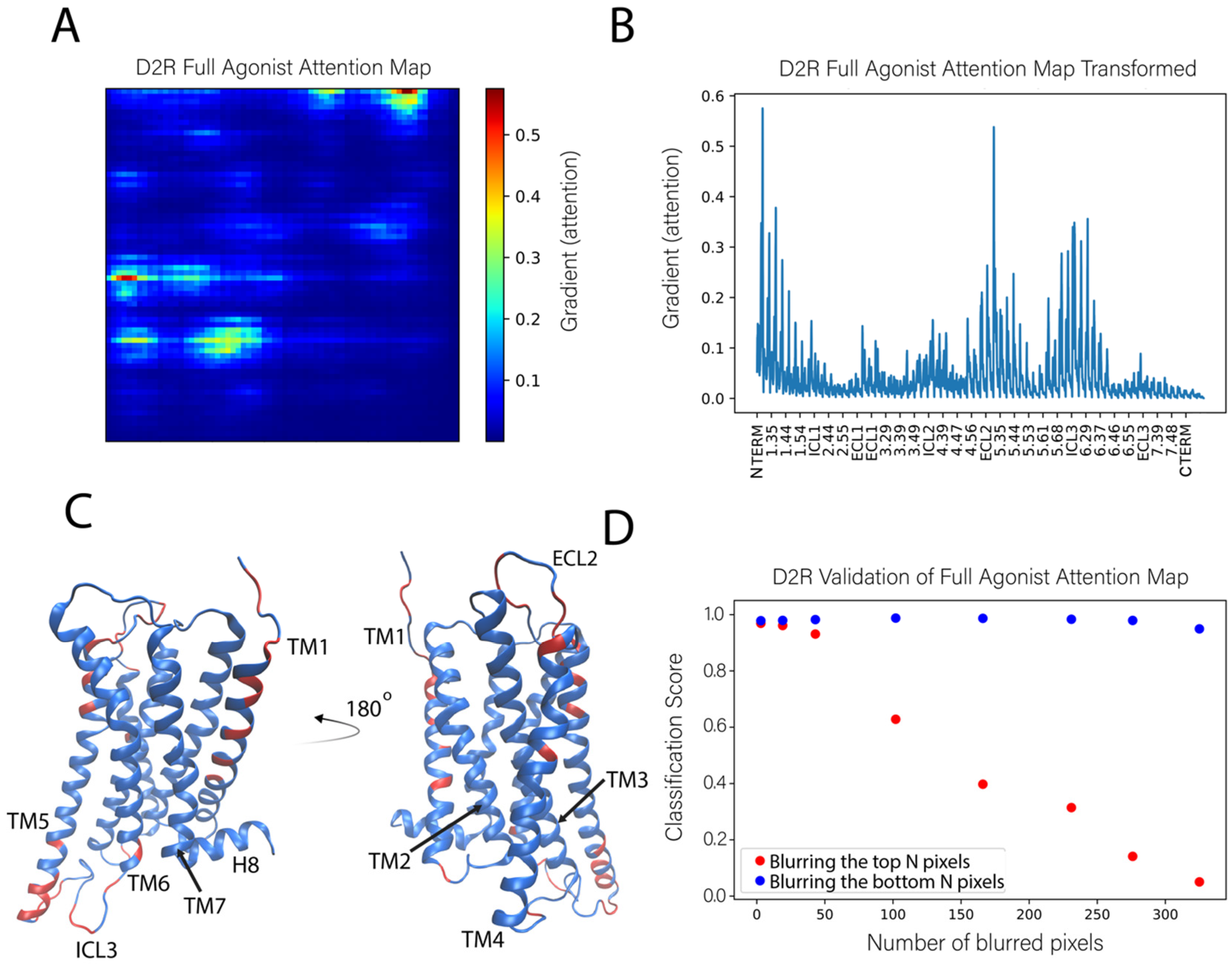
Molecules Free Full Text A Machine Learning Approach For The Discovery Of Ligand Specific Functional Mechanisms Of Gpcrs Html
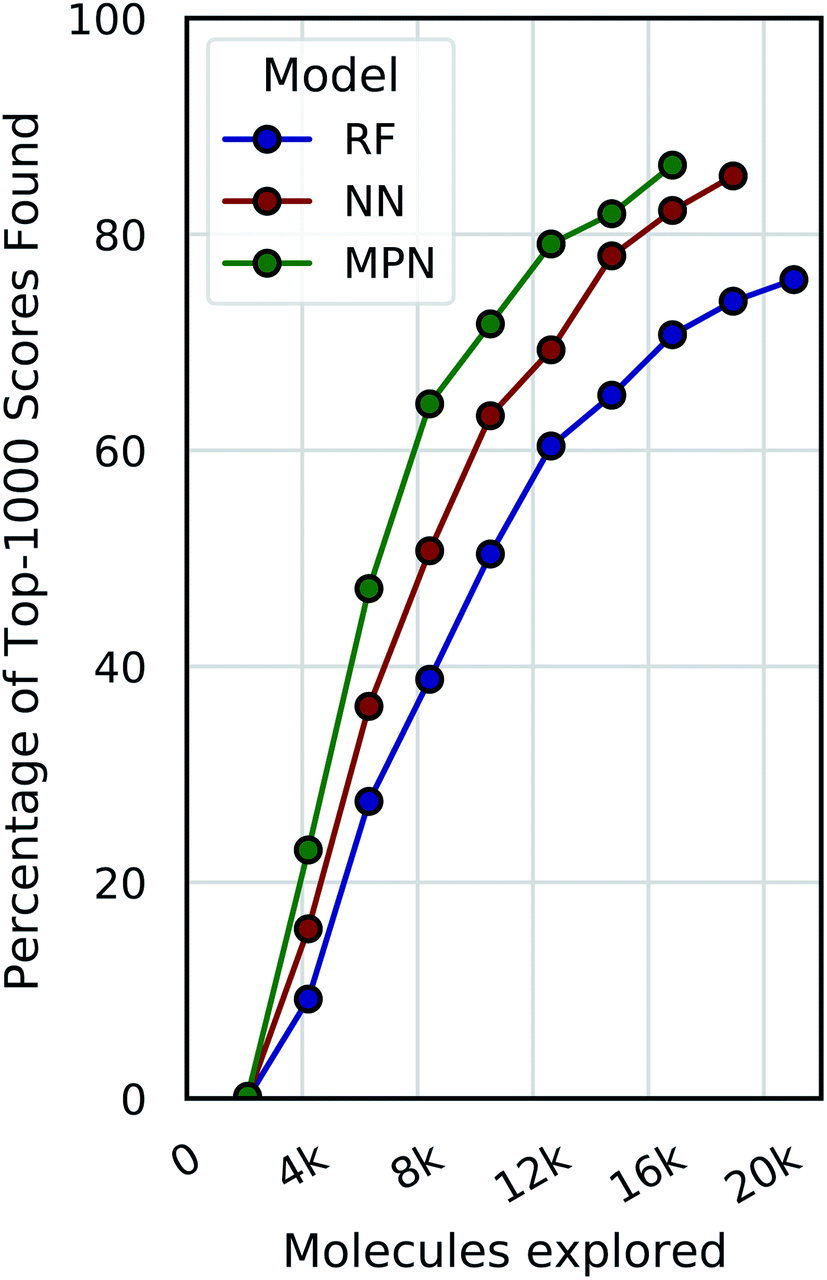
Accelerating High Throughput Virtual Screening Through Molecular Pool Based Active Learning Chemical Science Rsc Publishing Doi 10 1039 D0sc06805e

Application Advances Of Deep Learning Methods For De Novo Drug Design And Molecular Dynamics Simulation Bai Wires Computational Molecular Science Wiley Online Library
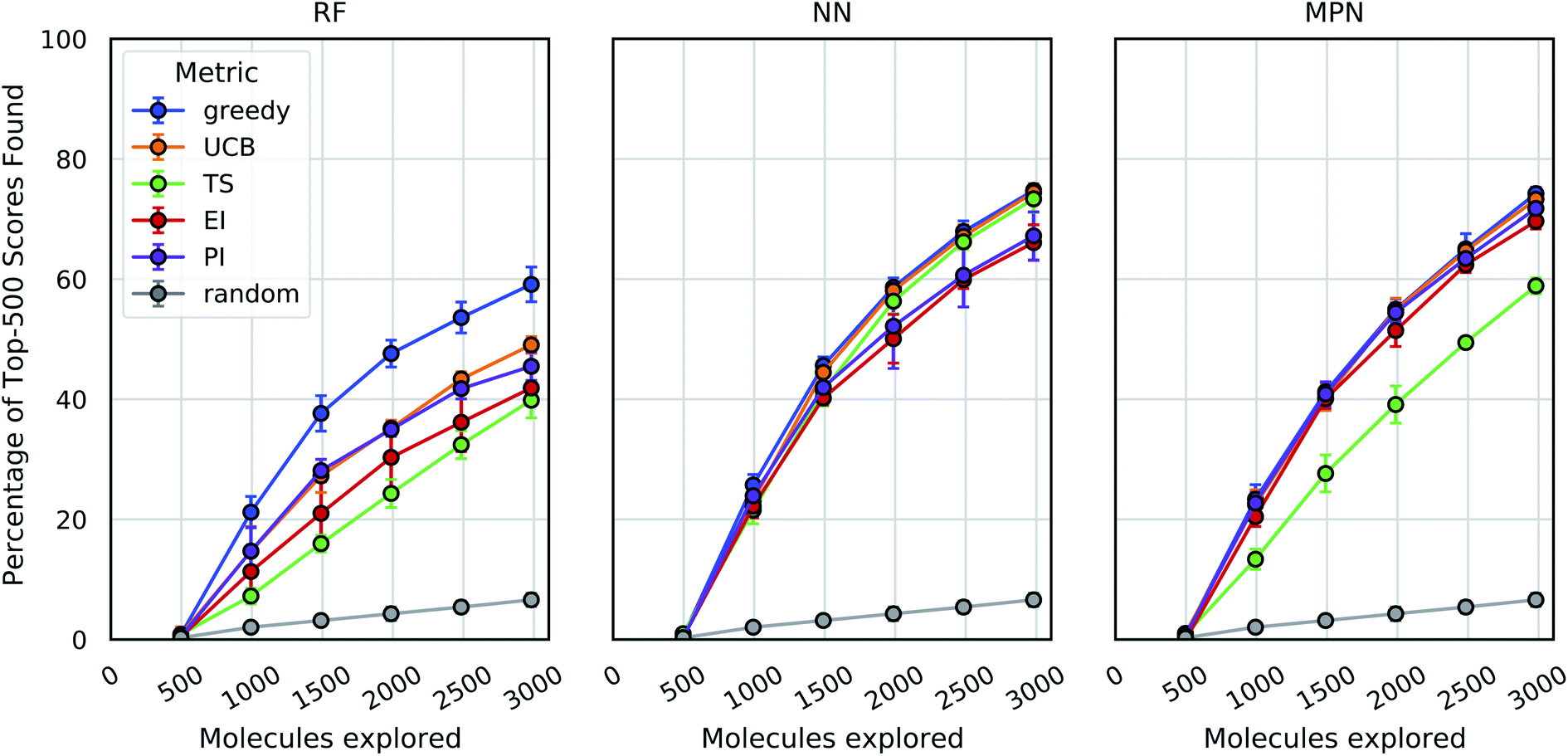
Accelerating High Throughput Virtual Screening Through Molecular Pool Based Active Learning Chemical Science Rsc Publishing Doi 10 1039 D0sc06805e

Pin On Ai Powered Drug Discovery

Schematic Representation Of A Deep Docking Dd Workflow Download Scientific Diagram
Komentar
Posting Komentar